
EI represents a preacylation noncovalent complex. Kinetic scheme of the reaction between a PBP (E) and a β-lactam (I). This affinity implicitly refers to the c 50, and should not be confused with the strength of a noncovalent interaction. Here, as in the literature in general, PBPs are said to be or to have high or low affinity for β-lactams. The value of c 50 is equal to the ratio k 3/( k 2/ K d). The inhibitory potency of a β-lactam for a PBP is given by the c 50, which is the antibiotic concentration resulting in the inhibition of half the PBP molecules at a steady state (when the acylation and deacylation reactions proceed at the same rate). The second-order rate constant k 2/ K d is the efficiency of acylation, which allows calculation of the overall acylation rate at a given antibiotic concentration. The constants k 2 and k 3 describe the acylation and deacylation rates, respectively. The following nomenclature will be used throughout this review. The rate described by k 3 is negligible on the time scale of a bacterial generation. EI * is hydrolyzed at the rate k 3 to regenerate E and an inactivated product P ( Fig. A noncovalent complex EI is formed between the enzyme E and the inhibitor I, with the dissociation constant K d, from which acylation proceeds to form the covalent complex EI * with the rate k 2. The following kinetic model describes the reaction of PBPs with β-lactams. This complex is hydrolyzed very slowly, thus effectively preventing further reactions. The active site serine attacks the carbonyl of the β-lactam ring, resulting in the opening of the ring and formation of a covalent acyl-enzyme complex. Β-Lactams mimic the d-Ala- d-Ala dipeptide in an elongated conformation, particularly regarding the distribution of three electrostatic-negative wells ( Fig. The regions of negative electrostatic potential are indicated by arcs. Structural similarity between β-lactams and the natural substrate of the PBPs. Such stem peptides are called ‘branched’.
B LACTAM DISSOCIATION CONSTANT PBP3 FREE
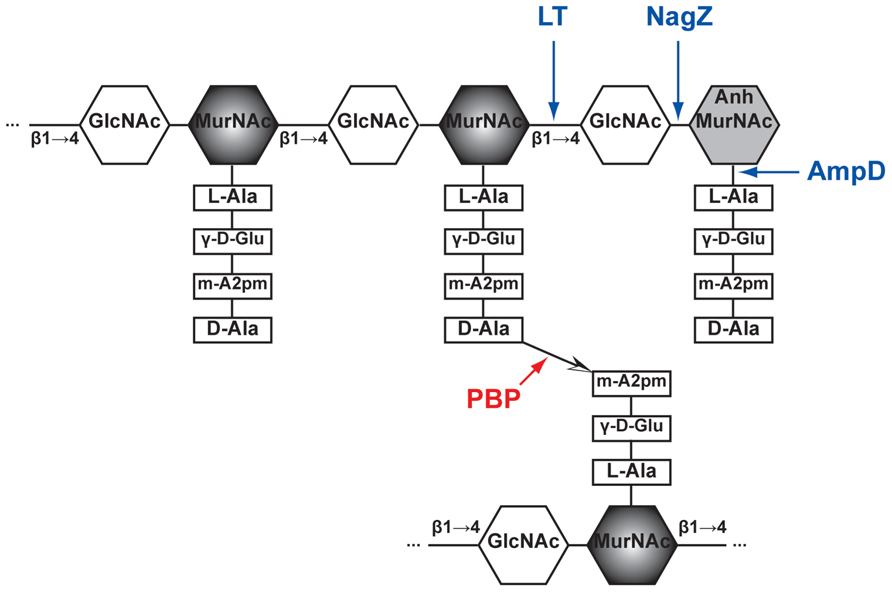
Note that in many instances, including in Streptococcus pneumoniae, various intervening amino acids are attached to the third residue of the acceptor peptide, and provide the free amine that attacks the acyl-enzyme intermediate. The second and third amino acids may differ in various species. The peptides shown are those from Streptococcus pneumoniae. The ‘donor’ pentapeptide is depicted on the upper glycan strand, whereas the ‘acceptor’ is attached here on the lower strand. Fragments of glycan strands are represented by chains of hexagones standing for the hexoses N-acetyl glucosamine (G) and N-acetyl muramic acid (M). In dd-carboxypeptidases, the acyl-enzyme intermediate is hydrolyzed this process eliminates ‘donor’ stem peptides from the peptidoglycan. A peptide bridge is then created between two stem peptides, forming a link between glycan strands ( Fig. In transpeptidases, the carbonyl of the d-Ala amino acid, now forming an ester linkage with the active site serine, then undergoes an attack from a primary amine linked in various ways to the third residue of a second ‘acceptor’ stem peptide. The serine attacks the carbonyl of the penultimate d-Ala amino acid of the stem peptide, which releases the last d-Ala amino acid from the ‘donor’ peptide and forms a covalent acyl–enzyme complex. The serine of the SXXK motif is central to the catalytic mechanism. These domains harbor three specific motifs: SXXK, (S/Y)XN and (K/H)(S/T)G, which define the active-site serine penicillin-recognizing enzymes family (for ASPRE) that also includes the class A and C β-lactamases. The penicillin-binding domains of PBPs are transpeptidases or carboxypeptidases involved in peptidoglycan metabolism ( Goffin & Ghuysen, 2002 Sauvage et al., 2008). Since their discovery as targets of the β-lactams, the penicillin-binding proteins (PBPs) have been the subject of intense research, particularly regarding their role in the resistance to β-lactams of some important pathogens such as Staphylococcus aureus, Enterococci and Streptococcus pneumoniae. Staphylococcus aureus, Streptococcus pneumoniae, Neisseria, Enterococcus What are PBPs?


 0 kommentar(er)
0 kommentar(er)
